Please Login to Use Workflows
| MassIVE Repository Statistics | |||
|---|---|---|---|
| Public Datasets: | Proteins: | ||
| Number of Files: | Peptides: | ||
| Total Size: | Peptide Variants: | ||
| Spectra: | PSMs: | ||
| Dataset Subscriptions: | Modifications: | ||
| Search Dataset Identifiers or Metadata: | |
| Search Universal Spectrum Identifier (USI): | |
| (example USI) |
MassIVE is a community resource developed by the NIH-funded
Center for Computational Mass Spectrometry to promote the
global, free exchange of mass spectrometry data.
MassIVE datasets can be assigned ProteomeXchange
accessions to satisfy publication requirements.
Supported by the National Institute of General Medical Sciences
of the National Institutes of Health under Award Number R24GM148372
Access Public Datasets |
Submit Data |
|---|---|
 Browse publically available datasets
or search by dataset metadata (e.g., species, PI,
etc.). Datasets are available for download as well
as for online browsing of submitted identifications
(for complete datasets). Dataset owners can also
add missing/requested files, update metadata and
add publications to their datasets. Registered
users can comment on datasets so others in the
community can see updates or find pointers to new
analyses of the data.
Browse publically available datasets
or search by dataset metadata (e.g., species, PI,
etc.). Datasets are available for download as well
as for online browsing of submitted identifications
(for complete datasets). Dataset owners can also
add missing/requested files, update metadata and
add publications to their datasets. Registered
users can comment on datasets so others in the
community can see updates or find pointers to new
analyses of the data.
|
 Submit your data
to share with the community as a MassIVE dataset;
reviewer access credentials
and
ProteomeXchange identifiers
can be requested to meet publication guidelines of
proteomics datasets
(documentation here).
Additional workflows are also available to
convert vendor raw files
(mass spectrometry data) to the open mzML format
and to
convert from common tab-separated formats
(identifications data) into the open mzTab format.
Submit your data
to share with the community as a MassIVE dataset;
reviewer access credentials
and
ProteomeXchange identifiers
can be requested to meet publication guidelines of
proteomics datasets
(documentation here).
Additional workflows are also available to
convert vendor raw files
(mass spectrometry data) to the open mzML format
and to
convert from common tab-separated formats
(identifications data) into the open mzTab format.
|
Search Identifications
|
Reanalyze Spectra |
 Search all submitted identifications
in complete datasets and dataset reanalyses. Over
300 million peptide-spectrum matches submitted with
at most 1% false discovery rate are accessible
through this simple interface to search for
peptides, proteins and post-translational
modifications.
Search all submitted identifications
in complete datasets and dataset reanalyses. Over
300 million peptide-spectrum matches submitted with
at most 1% false discovery rate are accessible
through this simple interface to search for
peptides, proteins and post-translational
modifications.
|
 Reanalyze public datasets
using online MassIVE workflows for analysis of mass
spectrometry data:
MSGF+
database search,
MSPLIT
spectral library search,
MODa
open modification search,
Maestro
spectral networks search and
MSPLIT-DIA
for search of data-independent acquisition (DIA)
spectra.
Reanalyze public datasets
using online MassIVE workflows for analysis of mass
spectrometry data:
MSGF+
database search,
MSPLIT
spectral library search,
MODa
open modification search,
Maestro
spectral networks search and
MSPLIT-DIA
for search of data-independent acquisition (DIA)
spectra.
|
Results Comparison |
Share Reanalyses |
 Compare identification results
between datasets or against any reanalyses of
public data. Venn diagrams are used to compare
results at the level of protein, peptide and
spectrum identifications; agreements, disagreements
and unique identifications can be interactively
inspected for assessment of quality of
identifications.
Compare identification results
between datasets or against any reanalyses of
public data. Venn diagrams are used to compare
results at the level of protein, peptide and
spectrum identifications; agreements, disagreements
and unique identifications can be interactively
inspected for assessment of quality of
identifications.
|
 Share your dataset reanalysis
results with the community: reveal new
identifications with novel algorithms / analysis
pipelines or challenge previously submitted
identifications with alternative interpretations
for the same data.
Share your dataset reanalysis
results with the community: reveal new
identifications with novel algorithms / analysis
pipelines or challenge previously submitted
identifications with alternative interpretations
for the same data.
|
Protein Explorer |
MassIVE Knowledge Base |
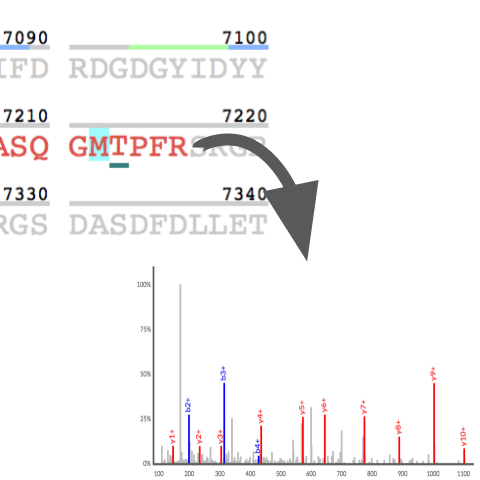 Explore
the translated evidence and sequence coverage of
nearly every human protein, as defined by
systematic reanalysis of 31 terabytes of public
data from >20,000 LC/MS runs and including over
1 million synthetic peptide spectra. Interactive
exploration of protein evidence includes coverage
maps, functional sites, and full provenance and
dataset mapping of every identified peptide.
Explore
the translated evidence and sequence coverage of
nearly every human protein, as defined by
systematic reanalysis of 31 terabytes of public
data from >20,000 LC/MS runs and including over
1 million synthetic peptide spectra. Interactive
exploration of protein evidence includes coverage
maps, functional sites, and full provenance and
dataset mapping of every identified peptide.
|
 Browse
the community big data derived MassIVE Knowledge
Base (MassIVE-KB) peptide spectral libraries.
Distilled from 31TB of human proteomics HCD data.
Users can peak at the inside of these libraries,
browse the source data, and track full provenance
of analysis tasks that created these libraries.
Please refer to the
documentation.
Browse
the community big data derived MassIVE Knowledge
Base (MassIVE-KB) peptide spectral libraries.
Distilled from 31TB of human proteomics HCD data.
Users can peak at the inside of these libraries,
browse the source data, and track full provenance
of analysis tasks that created these libraries.
Please refer to the
documentation.
|
MassIVE.quant |
CoronaMassKB |
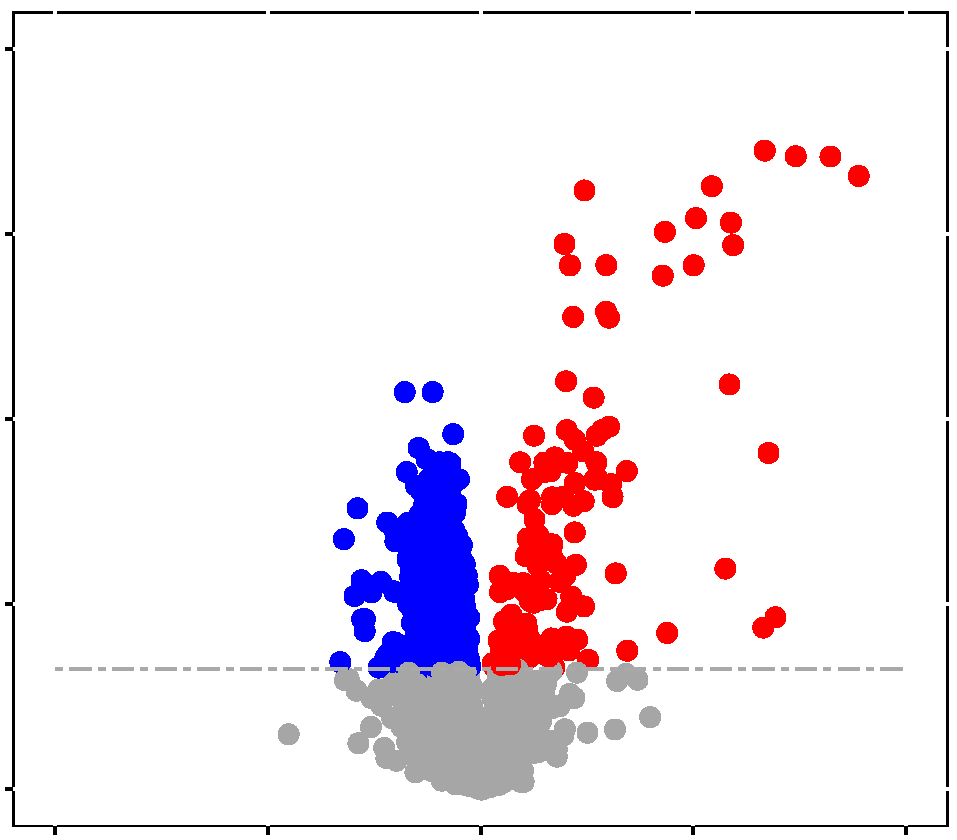 MassIVE.quant
is an extension of the Mass Spectrometry Interactive Virtual Environment
(MassIVE) to provide the opportunity for large-scale deposition of data
from quantitative mass spectrometry-based proteomic experiments.
MassIVE.quant is compatible with all mass spectrometry data acquisition
types and all computational analysis tools. For each dataset,
MassIVE.quant systematically stores the raw experimental data, the
annotations of the experimental design, the scripts (or descriptions) of
every step of the quantitative analysis workflow, and the intermediate
input and output files. A branch structure enables MassIVE.quant to store
and view alternative reanalyses of the same dataset with various
combinations of methods and tools in a way which allows the user to
inspect, reproduce or modify any component of the workflow, beginning
with well-defined intermediate files. MassIVE.quant supports
infrastructure to fully automate a analysis workflow, or to store, and to
browse the intermediate results.
MassIVE.quant
is an extension of the Mass Spectrometry Interactive Virtual Environment
(MassIVE) to provide the opportunity for large-scale deposition of data
from quantitative mass spectrometry-based proteomic experiments.
MassIVE.quant is compatible with all mass spectrometry data acquisition
types and all computational analysis tools. For each dataset,
MassIVE.quant systematically stores the raw experimental data, the
annotations of the experimental design, the scripts (or descriptions) of
every step of the quantitative analysis workflow, and the intermediate
input and output files. A branch structure enables MassIVE.quant to store
and view alternative reanalyses of the same dataset with various
combinations of methods and tools in a way which allows the user to
inspect, reproduce or modify any component of the workflow, beginning
with well-defined intermediate files. MassIVE.quant supports
infrastructure to fully automate a analysis workflow, or to store, and to
browse the intermediate results.
|
 CoronaMassKB
is an open-data community resource for sharing of mass spectrometry data
and (re)analysis results for all experiments pertinent to the global
SARS-CoV-2 pandemic. CoronaMassKB is designed for the rapid exchange of
data and results among the global community of scientists working towards
understanding the biology of SARS-CoV-2/COVID19 and thus accelerating the
emergence of effective responses to this global pandemic.
CoronaMassKB
is an open-data community resource for sharing of mass spectrometry data
and (re)analysis results for all experiments pertinent to the global
SARS-CoV-2 pandemic. CoronaMassKB is designed for the rapid exchange of
data and results among the global community of scientists working towards
understanding the biology of SARS-CoV-2/COVID19 and thus accelerating the
emergence of effective responses to this global pandemic.
|
Copyright © 2025.
Last modified: 2025-11-19.
Version 1.3.16-MassIVE.

